So, in my day job, I am a Senior AI Engineer at Lotus Health, the world’s first Primary Care Copilot aimed at patients(sign up for the waitlist if you’re interested!). There I spend a lot of time thinking about how to use LLMs to predict diseases or to do data processing across lots of medical data to tell you useful stuff. As I was working on Deepstock, I thought: hey, I should do something like this for disease prediction. And thus, I present: Diabetes Seek.
Basically, diabetes is a massive issue in the US. In the USA, according to the National Diabetes Statistics Report from the Centres for Disease Control and Prevention (CDC), 29·7 million people (including 29·4 million adults) were living with diabetes in 2021. Of those, 8·7 million (28%) of adults were undiagnosed, corresponding to 3·4% of all adults in the USA. 1
So, basically, the question is: Can we predict whether someone has diabetes or not, based solely on their existing conditions and medical history?
Essentially, my features for this would be:
a list of all the patient’s conditions prior to being diagnosed with diabetes if they have diabetes;
or just a list of conditions overall if they don’t.
The reasoning here is that pre-existing conditions are frequently a major risk factor for diabetes, and so could serve as a good basis for inferring whether or not someone has diabetes.
Data Preperation
I started with the MIMIC-IV database. The Medical Information Mart for Intensive Care (MIMIC)-IV database provides critical care data for over 299,712 patients admitted to intensive care units at the Beth Israel Deaconess Medical Center (BIDMC) between 2008 - 2019. Access can be granted at PhysioNet, it just takes a week or so.
From the MIMIC-IV database, I used DuckDB to join the diagnoses, admissions and definitions table to create a single all encompassing “clinical_data” table that gives a single large table that essentially contains the whole medical history for all the patients in the database. DuckDB is amazing as a data preprocessing tool: Super quick queries, and it autoloads pretty much any file format you want.
Here’s how the data looks(the example has been deidentified and scrambled):
admissions.csv, which shows the admissions for all patients.
diagnoses_icd.csv, which shows the diagnosises for each patient and admission.
and finally, the definitions table, d_icd_diagnoses.csv
Here is the code for converting the above into a clinical_data csv file which essentially joins all these tables together into one.
After this, I took the resulting clinical_data table, and for each patient, I created a set of features, which included:
Demographics
Complete Medical Timeline:
icd_code
icd_version
diagnosis_description
admittime_ts
Total number of admissions
(Note: I precalculate the timelines with one large select query which is subsequently grouped into a python dictionary keyed by patient_id)
Essentially, this creates patient histories that look like:
Patient History:
<Condition 1> - Year 0 Day 0
<Condition 2> - Year 1 Day 1
Based on this medical history, will this patient develop diabetes?
Your answer should look like the following
<think>reasoning about why the patient has diabetes
</think><answer>yes</answer>
Please reason about and provide several reasons for why you think the patient has diabetes in the <think></think> tags.
Please provide your answer as a single diagnosis, 'yes' or 'no', in the <answer></answer> tags, with yes meaning that the patient has diabetes and no meaning that the patient does not. Please put all the reasoning in the <think></think> tags, and don't write anything outside the <think></think> and <answer></answer> tagsThere are some nuances here:
Firstly, I provide all times as “Time in years and days since first admission”. This is since, due to deidentification, all the dates in the MIMIC-IV are shifted on by around a hundred years, so they all look like 01-09-2114. I didn’t want to bias the AI towards thinking this all takes place in the future, or any particular time for that matter, so I normalized all times into relative times.
Secondly, I filtered out all patients with less than 3 admissions, on the presumption that people with less wouldn’t provide enough good data to feed the AI. I checked and the resulting dataset also has around a 30% prevalence of diabetes, a bit higher than to the dataset prior to filtering, which has around 20% prevalence of diabetes, which is higher than population prevalence overall, although I chalked this up to people going to hospital being sicker than the norm.
This resulted in a fairly normalized database of about 56k rows, of which roughly a third have diabetes, all of which have a user prompt.
Training
For training, I utilized the GRPO Trainer implementation from Huggingface, building on their default training recipe in open-r1. I modified the configuration by increasing the completion length to approximately 1024 tokens while reducing the number of completions per step to 4.
My reward function focused on two aspects: First, a format reward that returns 1 if the answer follows the pattern `<think>arbitrary text</think> arbitrary text <answer> yes | no </answer>. I intentionally relaxed the format to permit arbitrary text between the </think> and <answer> tags after observing this behavior in early test runs, as I determined it wasn't detrimental to allow. Second, my accuracy reward was straightforward: 1.0 if the model's prediction correctly identified whether the user had diabetes, and 0.0 if the prediction was incorrect or failed to follow the required format.
I leveraged Modal for the training infrastructure due to its flexibility and scalability. Additionally, I made minor modifications to the trl implementation to output completions at each step, significantly improving visibility into the training process.
To run it, you save the above to “lotus_diabetes_seek.py” and the below to “main.py”, and then run `modal run main.py`
from modal import Image, App
import modal
huggingface_secret = modal.Secret.from_name(
"huggingface_secret", required_keys=["HF_TOKEN", "WANDB_API_KEY"]
)
# Create stub and image
app = App("diabetesseek")
GPU_USED = "A100-80GB:3"
DATASET_ID="lukasnellotus/clinical-all-patients-dataset-with-diabetes-labels-and-prompt"
MODEL_ID="deepseek-ai/DeepSeek-R1-Distill-Qwen-7B"
def download_models():
# Load model directly
from transformers import AutoTokenizer, AutoModelForCausalLM
from datasets import load_dataset
tokenizer = AutoTokenizer.from_pretrained(MODEL_ID)
model = AutoModelForCausalLM.from_pretrained(MODEL_ID)
dataset = load_dataset(DATASET_ID)
image = (
Image.debian_slim(python_version="3.11")
.pip_install("vllm==0.7.2", gpu=GPU_USED)
.apt_install("git")
.apt_install("wget")
.env({"HF_HUB_ENABLE_HF_TRANSFER": "1"}) # turn on faster downloads from HF
.run_commands("""ls && \
wget https://developer.download.nvidia.com/compute/cuda/repos/debian12/x86_64/cuda-keyring_1.1-1_all.deb && \
dpkg -i cuda-keyring_1.1-1_all.deb && \
apt-get update && \
apt-get install -y cuda-toolkit
""")
.run_commands(
'git clone https://github.com/huggingface/open-r1.git && cd open-r1 && pip install -e ".[dev]"', gpu=GPU_USED
)
.run_commands("ls && pwd")
.run_function(download_models, secrets=[huggingface_secret])
.pip_install("wandb")
.pip_install("peft")
#install fromm https://github.com/huggingface/trl/compare/main...LukasNel:trl:patch-2
.run_commands("pip uninstall -y trl && pip install git+https://github.com/LukasNel/trl.git@patch-2")
.add_local_file("lotus_diabetes_seek.py", "/open-r1/src/open_r1/grpo_lukas.py")
)
@app.function(image=image, secrets=[huggingface_secret], gpu="A100-80GB:4", timeout=43200,
volumes={
"/data": modal.Volume.from_name("diabetesseek-data")
})
async def run_training():
import os
import subprocess
os.chdir('/open-r1')
with open('zero3.yaml', 'w') as f:
f.write("""
compute_environment: LOCAL_MACHINE
debug: false
deepspeed_config:
deepspeed_multinode_launcher: standard
offload_optimizer_device: none
offload_param_device: none
zero3_init_flag: false
zero_stage: 2
distributed_type: DEEPSPEED
downcast_bf16: 'no'
machine_rank: 0
main_training_function: main
mixed_precision: bf16
num_machines: 1
num_processes: 4
rdzv_backend: static
same_network: true
tpu_env: []
tpu_use_cluster: false
tpu_use_sudo: false
use_cpu: false
""")
cmd = [
'accelerate', 'launch',
'--config_file', 'zero3.yaml',
'src/open_r1/grpo_lukas.py',
'--output_dir', '/data/DeepSeek-R1-Distill-Qwen-7B-GRPO-full',
# '--model_name_or_path', 'HuggingFaceTB/SmolLM2-1.7B-Instruct',
'--model_name_or_path',MODEL_ID,
"--report_to", "wandb",
'--dataset_name', DATASET_ID,
'--max_prompt_length', '256',
'--max_completion_length', '2048',
'--per_device_train_batch_size', '1',
'--gradient_accumulation_steps', '16',
"--save_steps", "10",
'--num_generations', '4',
"--log_completions", "True",
'--logging_steps', '1',
'--log_level', 'debug',
'--run_name', 'diabetesseek-check',
# '--project_name', 'diabetesseek',
# "--repo_id", "2084Collective/deepstock-v1",
'--num_train_epochs', '1',
'--bf16', 'true',
# "--use_peft", "true",
]
subprocess.run(cmd, check=True)
@app.local_entrypoint()
async def main():
await run_training.remote.aio()Results
I ran deepseek-ai/DeepSeek-R1-Distill-Qwen-7B for about 120 steps and got some interesting results.
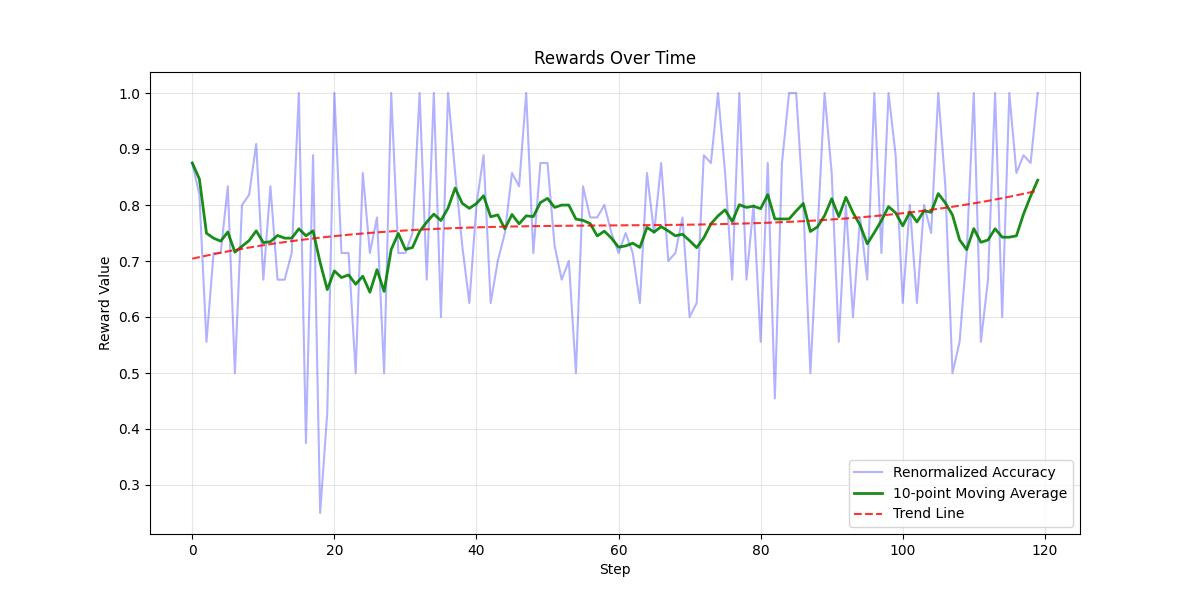
Since the accuracy reward is 0 for incorrect format, to get the actual accuracy, you have to divide the original accuracy by the format reward, as shown in the the following graph.
While the results look noisy, there are some interesting patterns:
Improved Accuracy: The average accuracy increased from around 70% to 85% over the whole training run, as shown by the 10-point moving average and trend line.
Performance Oscillation: The accuracy oscillates quite strongly between 1.0 (completely accurate) and 0.5 (completely unknown). Since a value of 0.5 means the model doesn't know whether or not the person has diabetes, that's actually a reasonable result: Sometimes, it's hard to tell from someone's history whether they have diabetes, and uncertainty is better than being completely wrong (which would be indicated by values below 0.5).
Learning Trajectory: The model went from occasionally dropping below 0.5 to mostly staying above it, which indicates it's definitely learning! There were also several instances where it achieved perfect accuracy (1.0).
This suggests that Diabetes Seek is a viable way to assess whether a person is likely to have diabetes: If the results consistently indicate "yes," then a person probably has diabetes; if it's 50-50, then the result is inconclusive—which is still better than being wrong.
As for user experience, the model's explanations in the <think></think> section make it significantly more user-friendly than most black-box models. Plus, this only inferred from a patient's medical history, with no blood tests included—if we had included those, the predictions would likely have been even more accurate.
Here’s an example of a patient report:
After reviewing the patient's medical history, several factors suggest a likelihood of developing diabetes. First, the patient has a history of hypertension, which is a known contributor to the development of type 2 diabetes. High blood pressure can lead to insulin resistance and eventual insulin resistance, contributing to the development of diabetes.
Second, the patient has significant hyperlipidemia, particularly elevated LDL cholesterol and triglycerides. High cholesterol levels are associated with insulin resistance and poor glucose metabolism, which can result in type 2 diabetes.
Third, the patient has a history of transient ischemic attack (TIA) and cerebral infarction. While these conditions are associated with stroke risk, they can also lead to changes in blood sugar levels, particularly hyperglycemia, which is a hallmark of insulin resistance and eventually type 2 diabetes.
Fourth, the patient has long-term use of anticoagulants, aspirin, and has had a surgical operation with an implant causing complications. While anticoagulants and aspirin are antiplatelet medications, they can sometimes have metabolic side effects such as increased glucose production, leading to hyperglycemia.
Additionally, the patient's current management of hypertension and hyperlipidemia without diabetes could lead to progression toward metabolic insulin resistance, especially if the current medications are not sufficient to control these conditions. The continued presence of glucose monitors and lab studies for HbA1c and fasting glucose suggests that controlling these conditions is a priority, but without intervention to address insulin resistance or insufficient antiplatelet therapy, this progression towards diabetes remains likely.
</think>
<answer>Yes</answer>
Reasoning: The patient has a history of hypertension, hyperlipidemia, a stroke risk (TIA and cerebral infarction), and complications from an implant device. These factors increase the likelihood of developing type 2 diabetes. Current management includes glucose monitoring and lab studies, which support the need for further diabetes control.Limitations and Future Work
This approach isn't entirely ready for prime time: The model hallucinates quite a bit in its reasoning reports, sometimes making up details about cholesterol and glucose levels despite that information not being provided. This is an inevitable consequence of using a relatively small model. With a larger model, these hallucinations would likely be reduced.
Potential Extensions
Include bloodwork results for more accurate diabetes inference
Apply the approach to other diseases
Use a much larger model
Train a separate model using traditional machine learning techniques to compare performance
Note on DeepStock
I've been training DeepStock on and off for the last couple of weeks, but the training keeps crashing with an out-of-memory error after only a few steps, although it always starts promisingly! My suspicion is that some of the prompts in the dataset are quite long, making the GRPO Trainer prone to running out of memory. I'll need to either rewrite the GRPO training code to be more memory efficient or dive deeper into how distributed training interacts with Huggingface's Trainer code, which I plan to do over the next week or so. I also want to use a larger model, which further necessitates better distributed training capabilities.







Fascinating how you formulated the problem. I can see lot of use if this in actuarial analyses of large sets of data, where possibly being right about every individual outcome is not as important as highlighting interesting subgrouos within the data set with high risk of, in this case, diabetes.